

I am a doctoral candidate in Dr. Lawrence David's laboratory in the Department of Molecular Genetics and Microbiology at Duke University. As a microbiologist, I am very interested in microbial interactions, both at the cellular level and within complex microbial ecosystems. My research has taken me through investigations of how bacteria kill one another and how dietary patterns influence the bacteria living in our gut. Over the past nine years, I have dedicated approximately 60% of my time to wet-lab experiments and 40% to computational data analysis and visualization.
I am also exploring how the integration of tools such as AI agents, MCPs, automation workflows, and vibe coding could streamline my project management and data analysis. I view AI tools as accelerators of learning and efficiency in research, though I do not support or utilize generative art. All figures and diagrams in my work are created by me manually using R or BioRender.
You can reach out to me via email or LinkedIn. A complete list of my publications is available on Google Scholar, and the code supporting my analyses and visualizations can be found on GitHub.
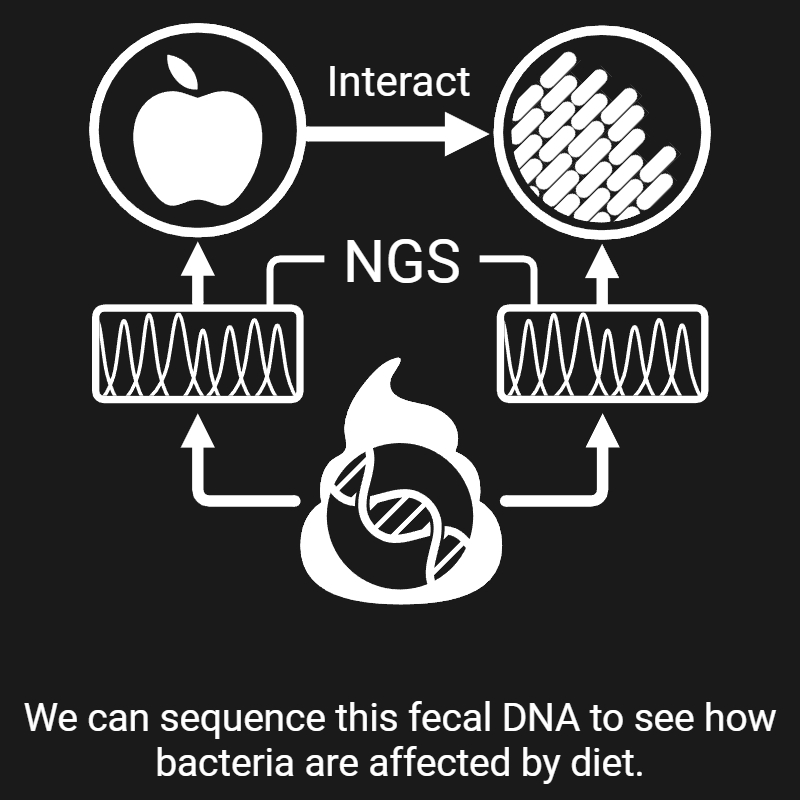
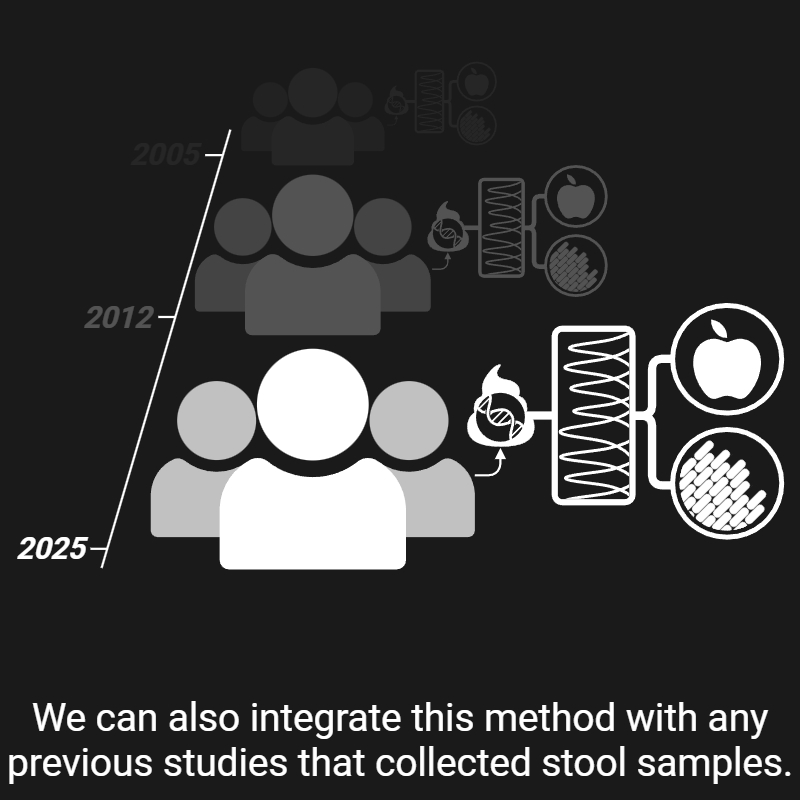
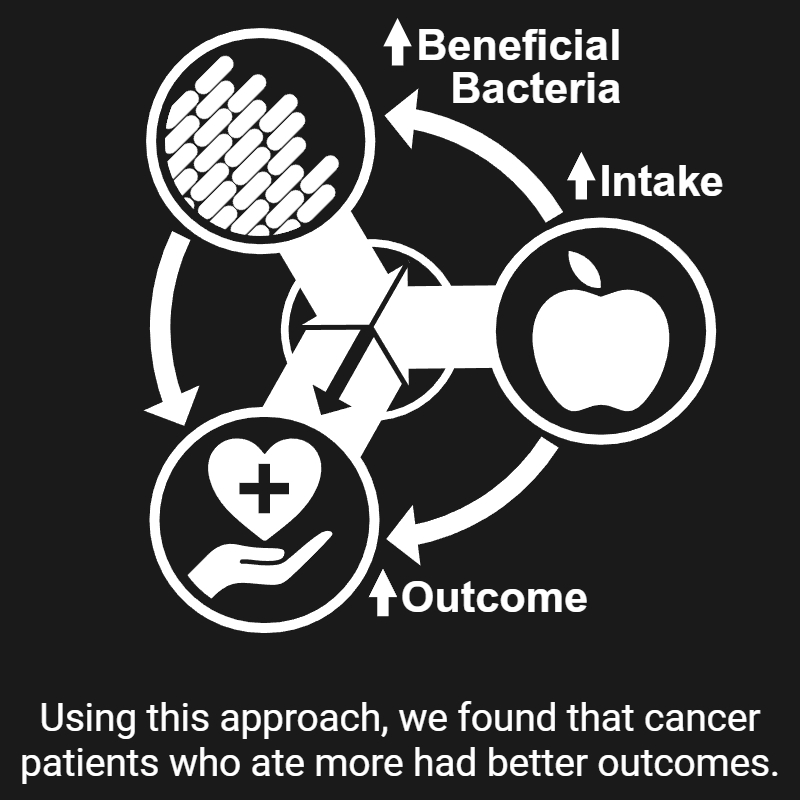
My research revolves around two central questions: 1) how microbes interact with the different aspects of their environment, and 2) how we can exploit these interactions to improve human health. Over the past nine years, I have been chasing answers to these questions—and here's what I've found, so far:




My experiences have provided me with a broad range of both experimental (wet lab) and computational (dry lab) skills, a list of which is provided on the right. Skills or tools that I used but did not end up in published works are indicated in light gray.
Zeng J., García-González A. P., Epstein P., Bauer A. E., Jiang S., Kirtley M. C., Neubert B. C., Rivera C. N., Bergens M. A., Bush A. T., Hill L., Gauthier J., McGriff C., Tang H., Andermann T. M., Jobin C., Chao N. J., Dahl W. J., Wingard J. R., Sung A. D., & David L. A. (2025). Dietary signatures from fecal DNA predict hematopoietic stem cell transplantation outcomes. Submitted.
Aqeel A., Kay M. C., Zeng J., Petrone B. L., Yang C., Truong T., Brown C. B., Jiang S., Carrion V. M., Bryant S., Kirtley M. C., Neshteruk C. D., Armstrong S. C., & David L. A. (2025). Grocery intervention and DNA-based assessment to improve diet quality in pediatric obesity: a pilot randomized controlled study. Obesity (Silver Spring, Md.), 33(2), 331–345. [Link]
Letourneau J., Carrion V. M., Zeng J., Jiang S., Osborne O. W., Holmes Z. C., Fox A., Epstein P., Tan C. Y., Kirtley M., Surana N. K., & David L. A. (2024). Interplay between particle size and microbial ecology in the gut microbiome. The ISME journal, 18(1), wrae168. [Link]
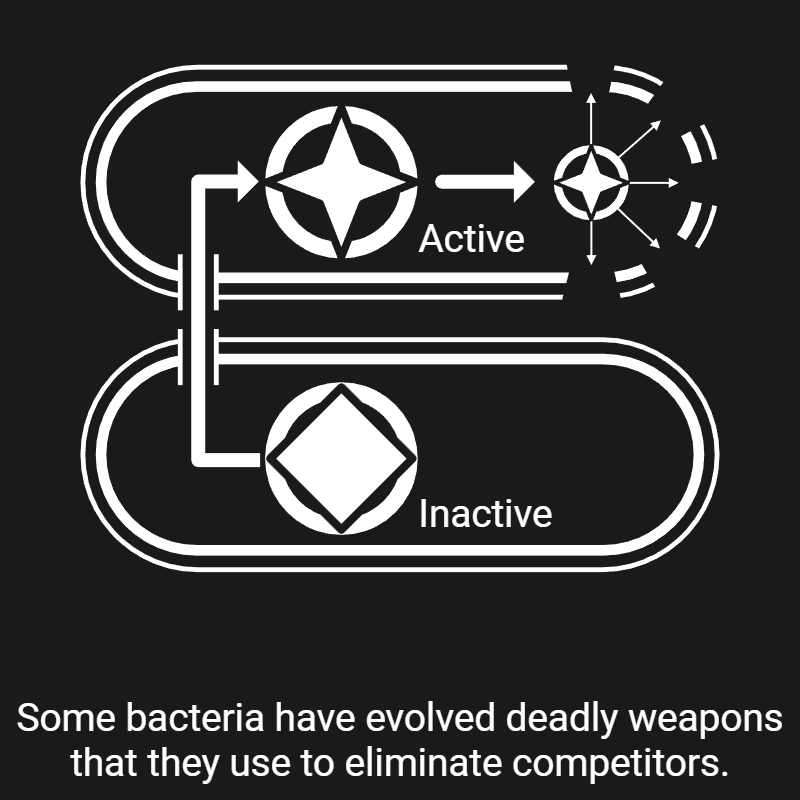
de Moraes M. H., Hsu F., Huang D., Bosch D. E., Zeng J., Radey M. C., Simon N., Ledvina H. E., Frick J. P., Wiggins P. A., Peterson S. B., & Mougous J. D. (2021). An interbacterial DNA deaminase toxin directly mutagenizes surviving target populations. eLife, 10, e62967. [Link]
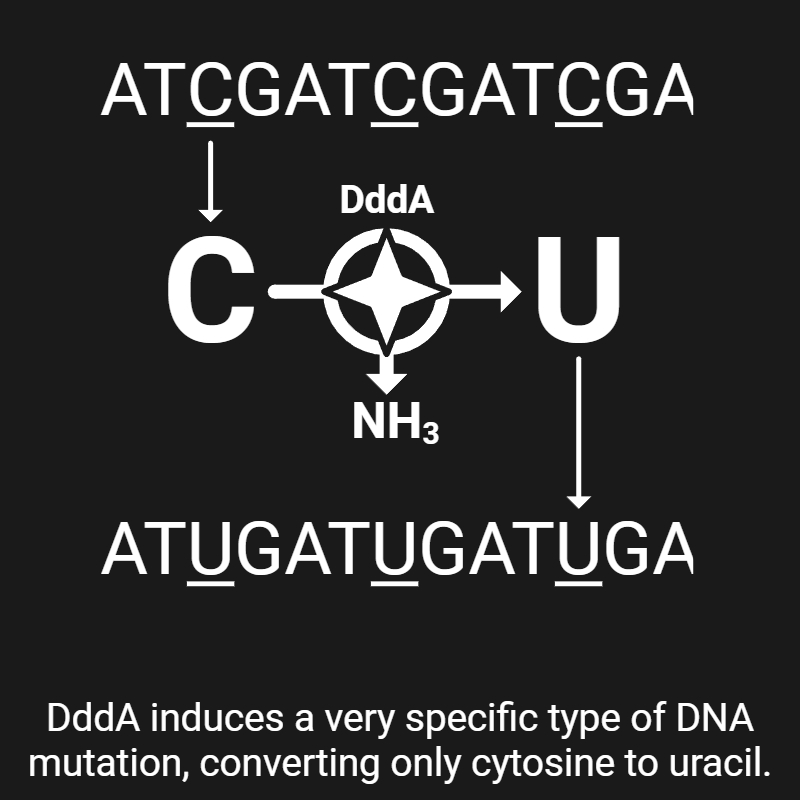
Mok B. Y., de Moraes M. H., Zeng J., Bosch D. E., Kotrys A. V., Raguram A., Hsu F., Radey M. C., Peterson S. B., Mootha V. K., Mougous J. D., & Liu D. R. (2020). A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature, 583(7817), 631–637. [Link]
Some examples of plots I made for my published works.
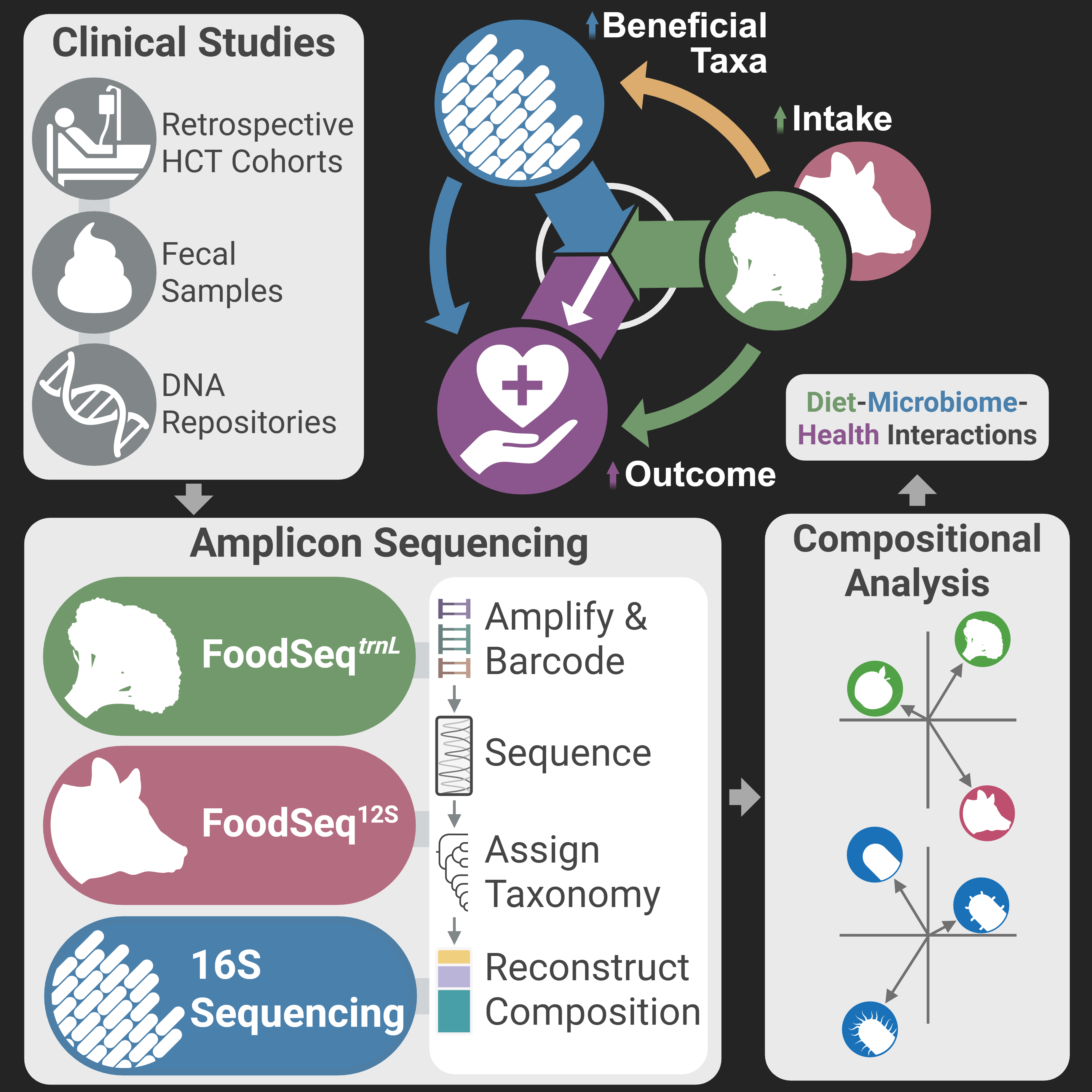
Graphical summary of my thesis project, which integrate our findings from amplicon sequencing of plant trnL, animal 12S, and bacterial 16S markers--all from the same DNA extracts originated from only 15 mg of the origial fecal materials.
Each data point is a fecal sample colored by week of sample collection relative to HCT. Double click on keys in the legend to filter samples.
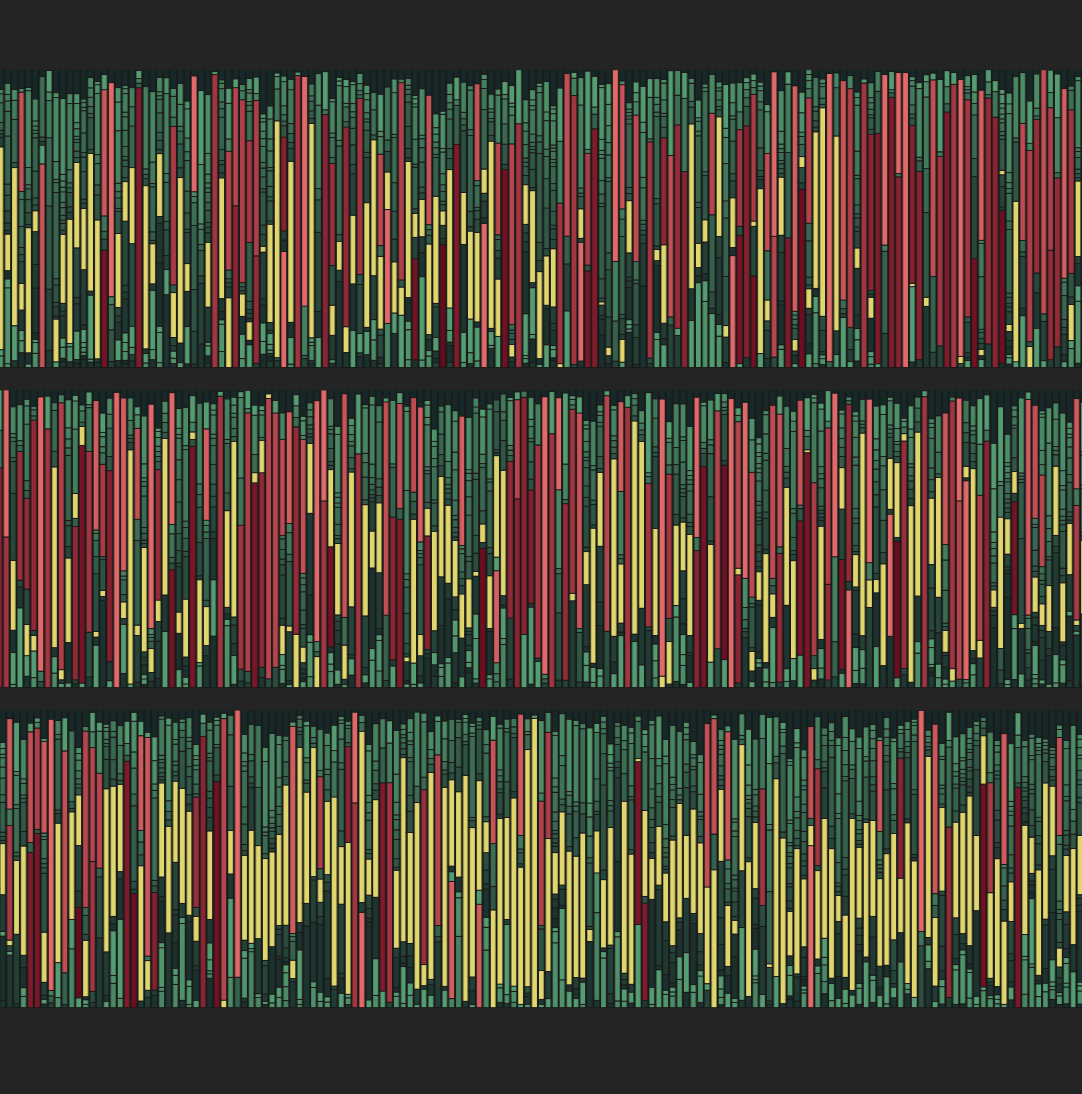
Each bar is microbiome composition of a stool sample, derived from 16S rRNA sequencing. Yellow indicates Blautia, red indicates non-Blautia species present at ≥30% relative abundance, and green indicates species present at <30% relative abundance.
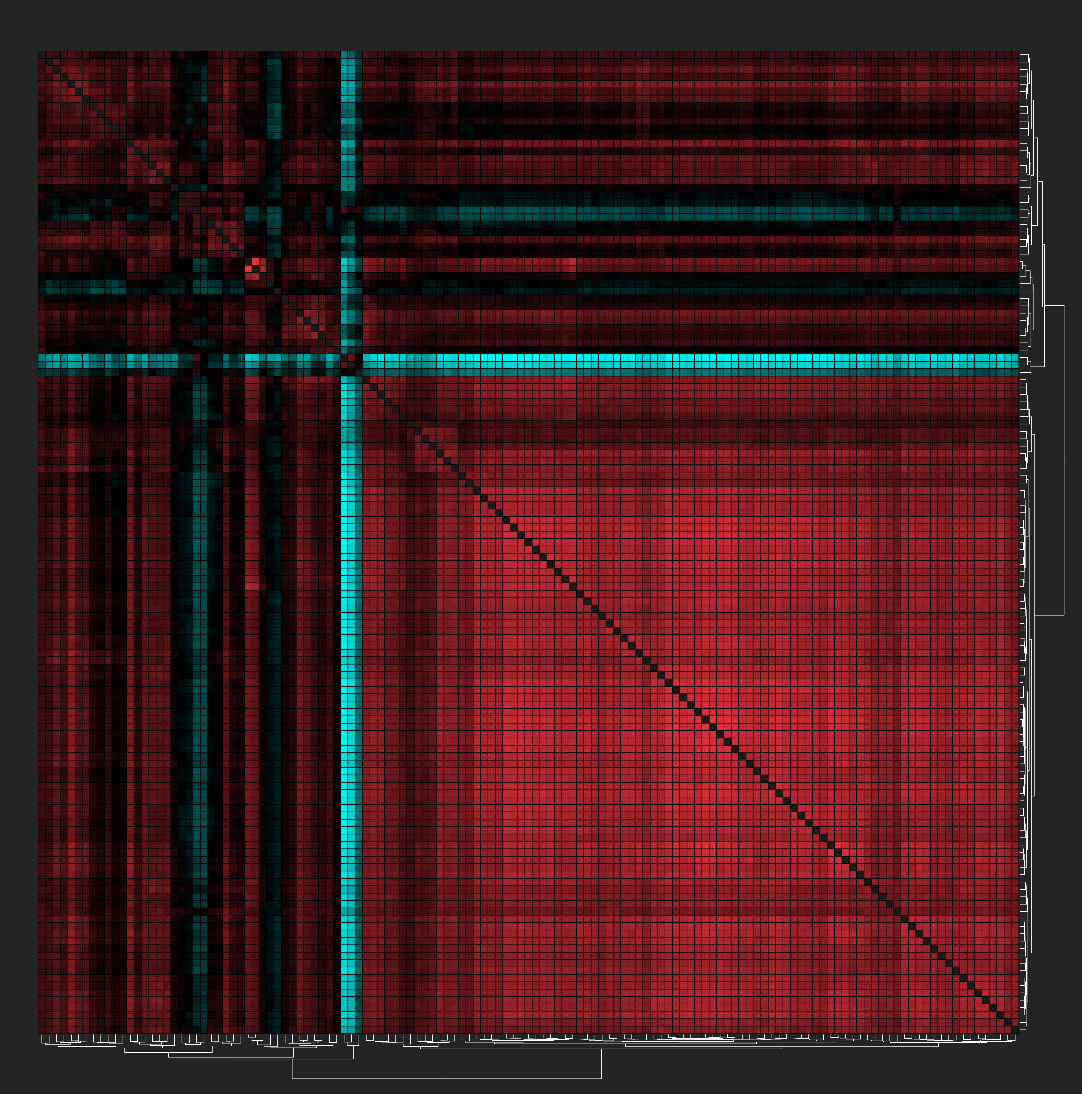
A heatmap displaying a Euclidean distance matrix based on bacterial abundance. Red indicates lower distances, whereas cyan represents higher distances. The three dominant microbiome taxa showed negative associations with almost all other microbial groups.
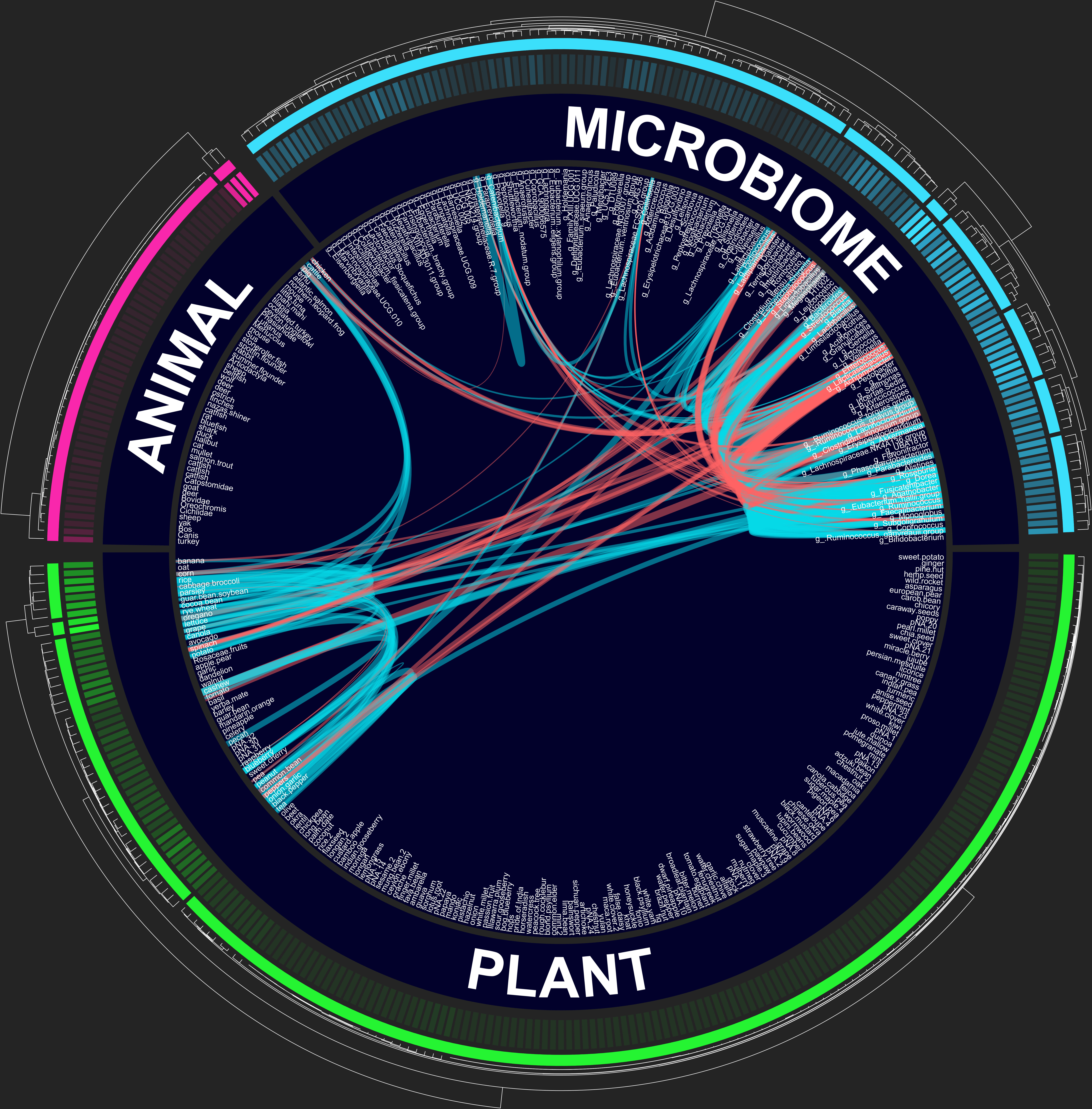
Associations are more apparent between food taxa or between bacterial taxa, while inter-dataset associations are rare. This is likely due to the fact that diet is more transient and dynamic, while bacterial compositions could be stable over longer periods of time.